Your mutation of interest in our opti-ox powered human iPSC-derived cells


With this custom offering, you can now order your mutation of choice in bit.bio’s opti-ox powered human-iPSC derived cells and receive consistent, defined, cryopreserved disease models that can be immediately incorporated into your experiments.
Every custom disease model comes with a genetically matched control, ioWild Type Cells, giving you confidence that even subtle variances in your data are attributable to your mutation of interest.
Remove the challenges associated with animal models, patient-derived cells and directed differentiation protocols from your workflows, and start collecting disease-relevant data you can trust in a human context. Each project begins as a conversation with our experts about your custom disease model needs.
Build your custom disease model into these wild type cell backgrounds. Use the genetically matched control in your experiments to make true comparisons in your data, and confidently link genotype to phenotype.
"The access to better human cell-based in vitro disease models as generated within our partnership is transformational for the way we do drug discovery with our pharma and biotech clients. We can now provide discovery studies in human stem cell-based assays, where previously we would only do this in for example immortalised cell lines or more simple assay types.”

Marijn Vlaming, PhD
Charles River Laboratories
Identifying intrinsic, observable phenotypes in ioDisease Model Cells has the potential to streamline compound testing in drug discovery by reducing the need to induce disease-related phenotypes when modelling specific diseases. Discover the data generated by our industry partners below.
Our disease models of tau dysfunction show a tau hyperphosphorylation phenotype important to frontotemporal dementia research.
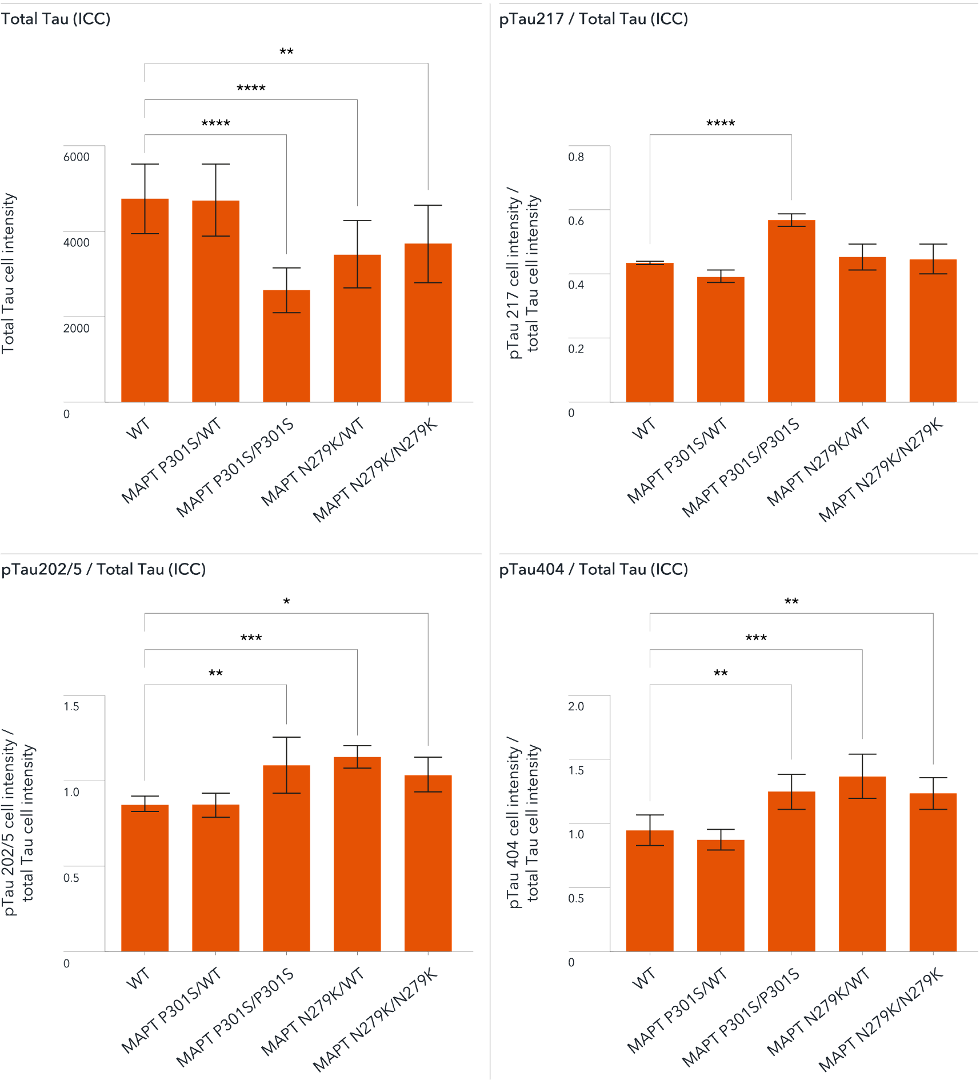
A panel of disease models for studying frontotemporal dementia (FTD) was developed by engineering either FTD-causing heterozygous or homozygous N279K or P301S mutations in the MAPT gene of ioGlutamatergic Neurons.
At day 21 post-revival, the data showed hyperphosphorylation of mutant tau protein compared to the wild type control using high-content image analysis. The panel offers the potential to investigate the impact of mutant tau on the molecular mechanisms of disease and test potential drug candidates rapidly in high throughput multiwell plate format. Data courtesy of Charles River Laboratories.
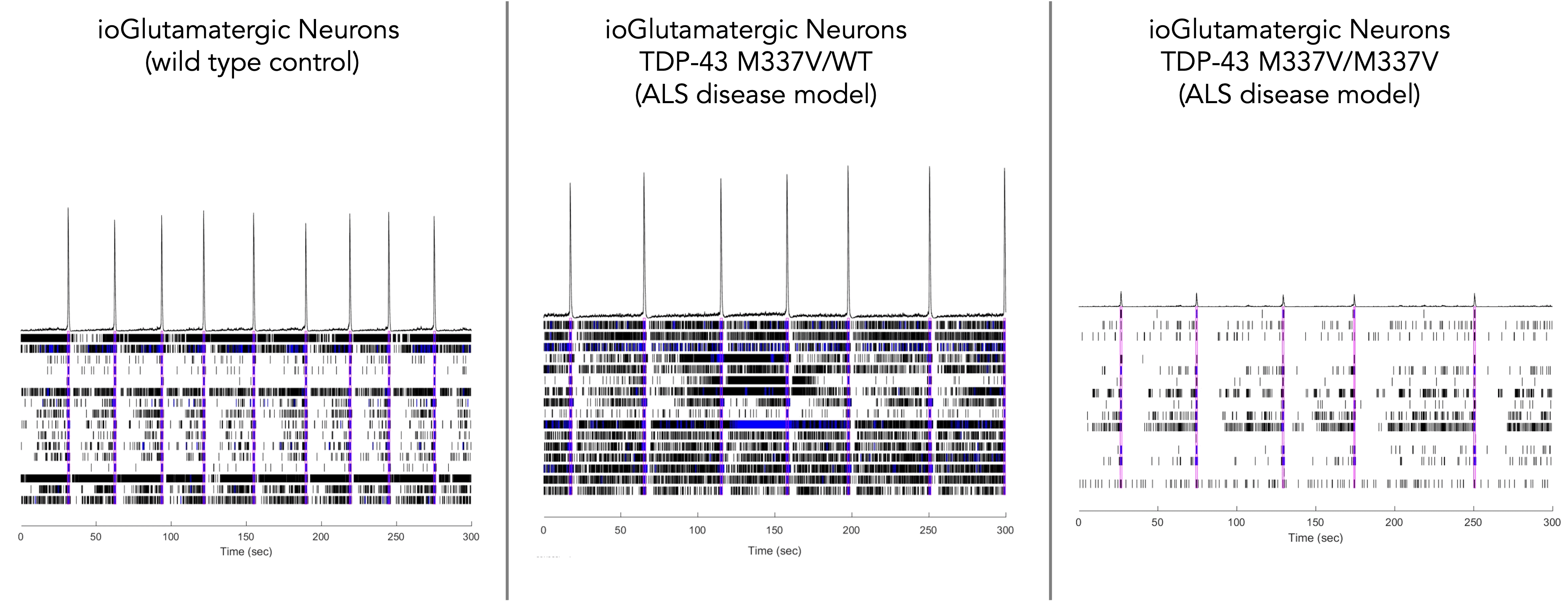
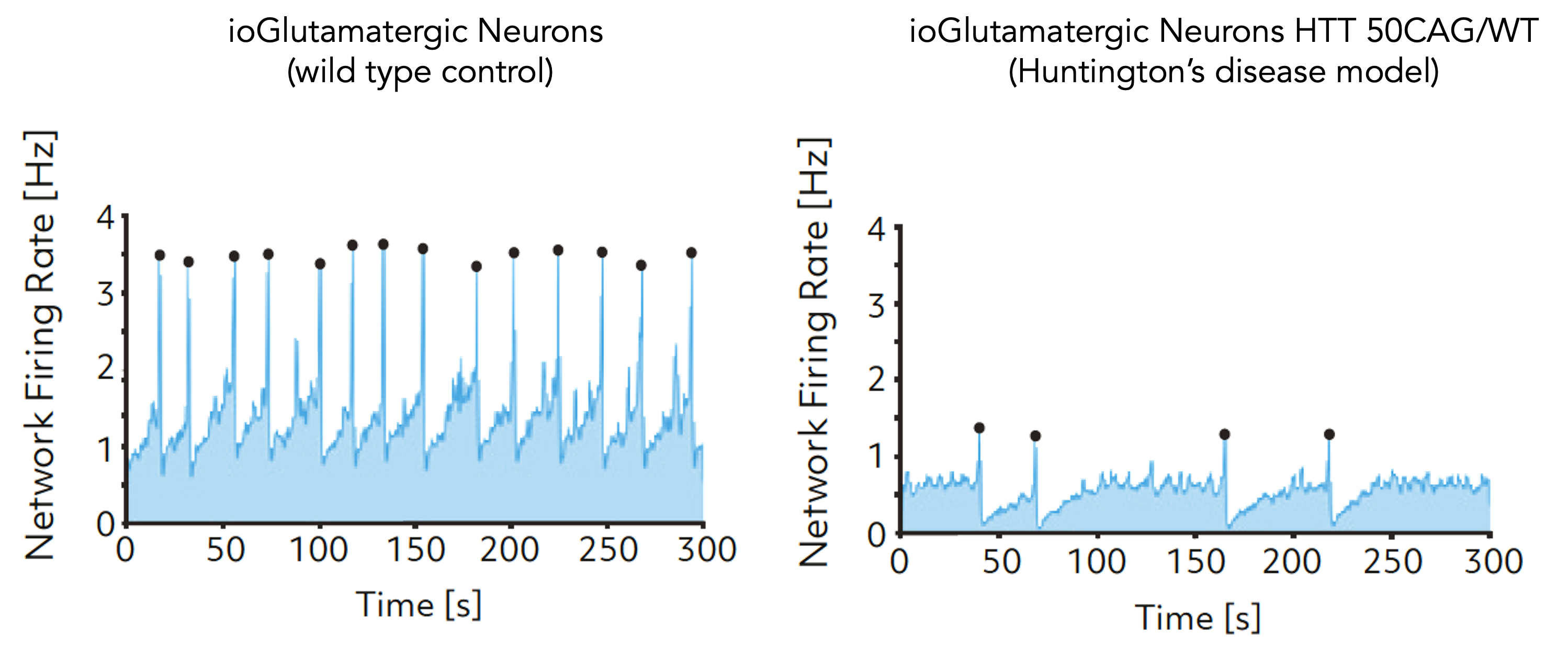
Using a microelectrode array (MEA)-based assay our ALS disease model (ioGlutamatergic Neurons TDP-43 M337V/M337V=) demonstrates a significant reduction in neuronal activity compared to the ALS disease model with a heterozygous mutation (ioGlutamatergic Neurons TDP-43 M337V/WT) and the wild-type isogenic control. Data courtesy of Charles River Laboratories.
This indicates their potential as a relevant translational in vitro drug discovery model for ALS.
Charles River Laboratories Discovery team now offers these disease models alongside high-content imaging and gene expression analysis to support investigations into TDP-43 protein phosphorylation, mislocalisation, aggregation, and altered mRNA splicing, which are hallmarks of ALS and FTD.
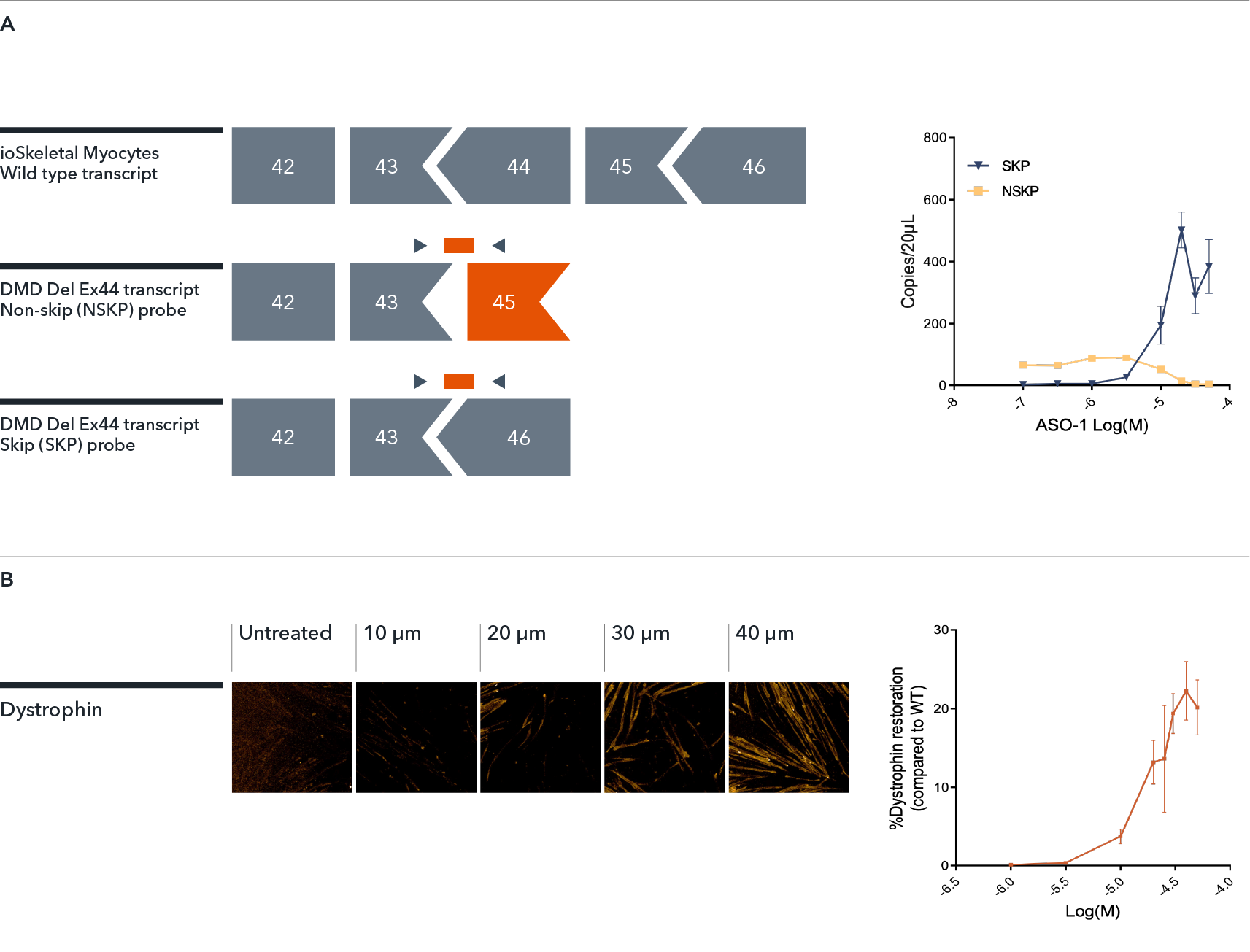
The DMD disease model offers a valuable translational model to investigate methods for dystrophin restoration, such as ASO-mediated exon skipping.
In this example, ioSkeletal Myocytes (wild type control) and ioSkeletal Myocytes DMD Exon 44 Deletion (DMD Del Ex44) were treated by gymnosis with exon 45 skipping antisense oligonucleotide (ASO-1). The data shows a concentration-dependent increase in the amount of mRNA transcript (A) and dystrophin protein (B) in the DMD exon 44 deletion disease model cells, indicating that ASO-1 treatment has been successful in creating an in frame mRNA transcript for dystrophin that leads to protein expression. Data courtesy of Charles River Laboratories.
Start a no-commitment project consultation with our experts today.

Consistent. Defined. Scalable.