Modelling neurodegeneration using a human isogenic system: A next generation approach to study frontotemporal dementia and amyotrophic lateral sclerosis
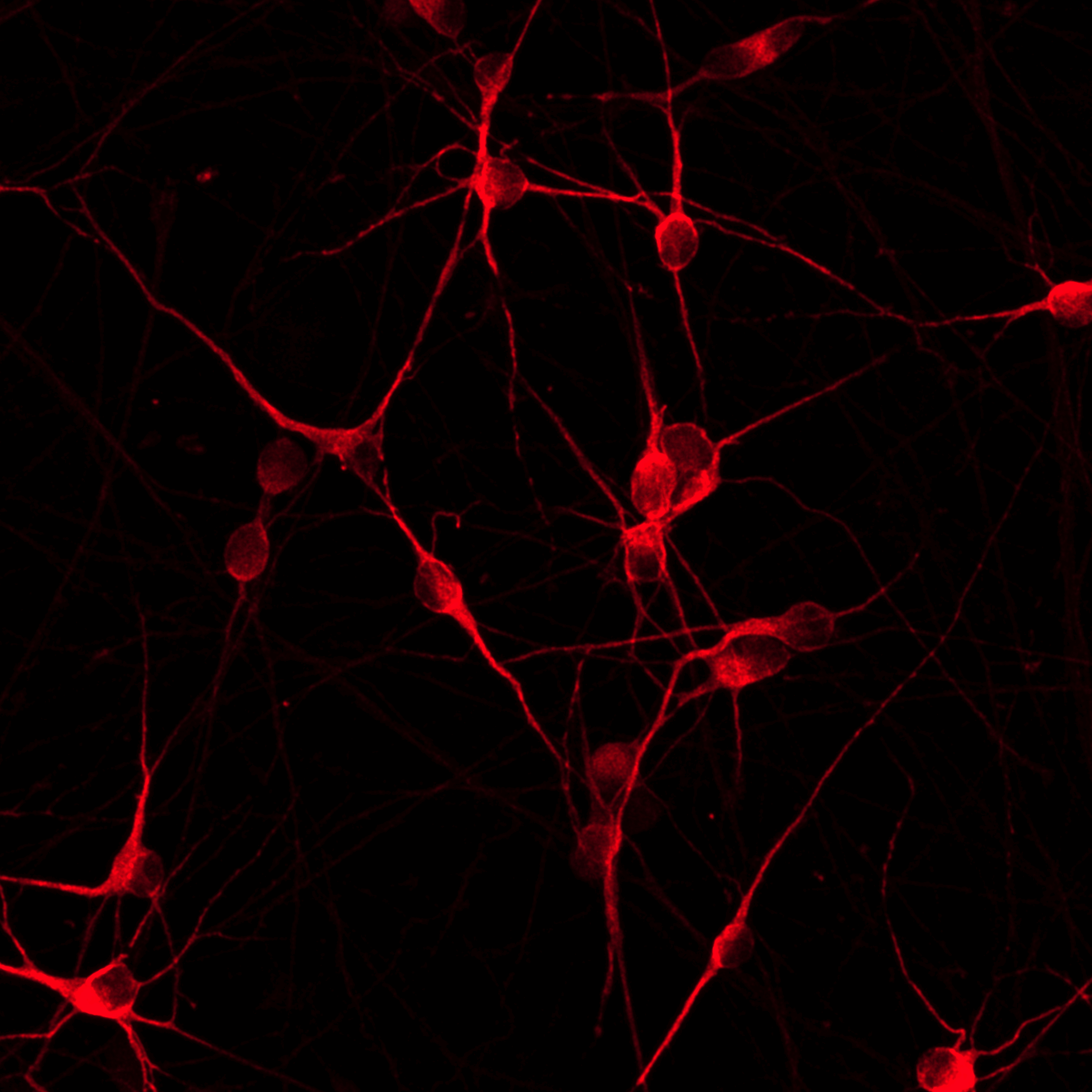
We have developed human iPSC-derived disease models for frontotemporal dementia (FTD) and amyotrophic lateral sclerosis (ALS) to improve screening specificity and accelerate early drug discovery for these neurodegenerative disorders. Using CRISPR/Cas9 gene editing, we introduced the disease-relevant mutation M337V in the TDP-43 (TARDBP) gene, or the mutation P301S or N279K in the MAPT gene, of wild type ioGlutamatergic Neurons (isogenic control). Disease-related phenotypic differences between the disease model cells and the isogenic control were detected by microelectrode array (MEA) analysis and high content immunocytochemistry image analysis.
In this poster, you will explore:
- How ioGlutamatergic Neurons carrying the TDP-43 M337V, MAPT N279K or MAPT P301S mutations were generated and characterised.
- Development of high content image (HCI) analysis and droplet digital PCR assays to compare the TDP-43 M337V disease models and isogenic control.
- MEA analysis showing a phenotype of reduced neuronal activity in the TDP-43 M337V homozygous disease model compared to the isogenic control.
- Demonstration of a tau hyperphosphorylation phenotype in the MAPT P301S and N279K disease models by HCI analysis.
The data demonstrate the successful generation of relevant translational in vitro drug discovery models for ALS and FTD.
