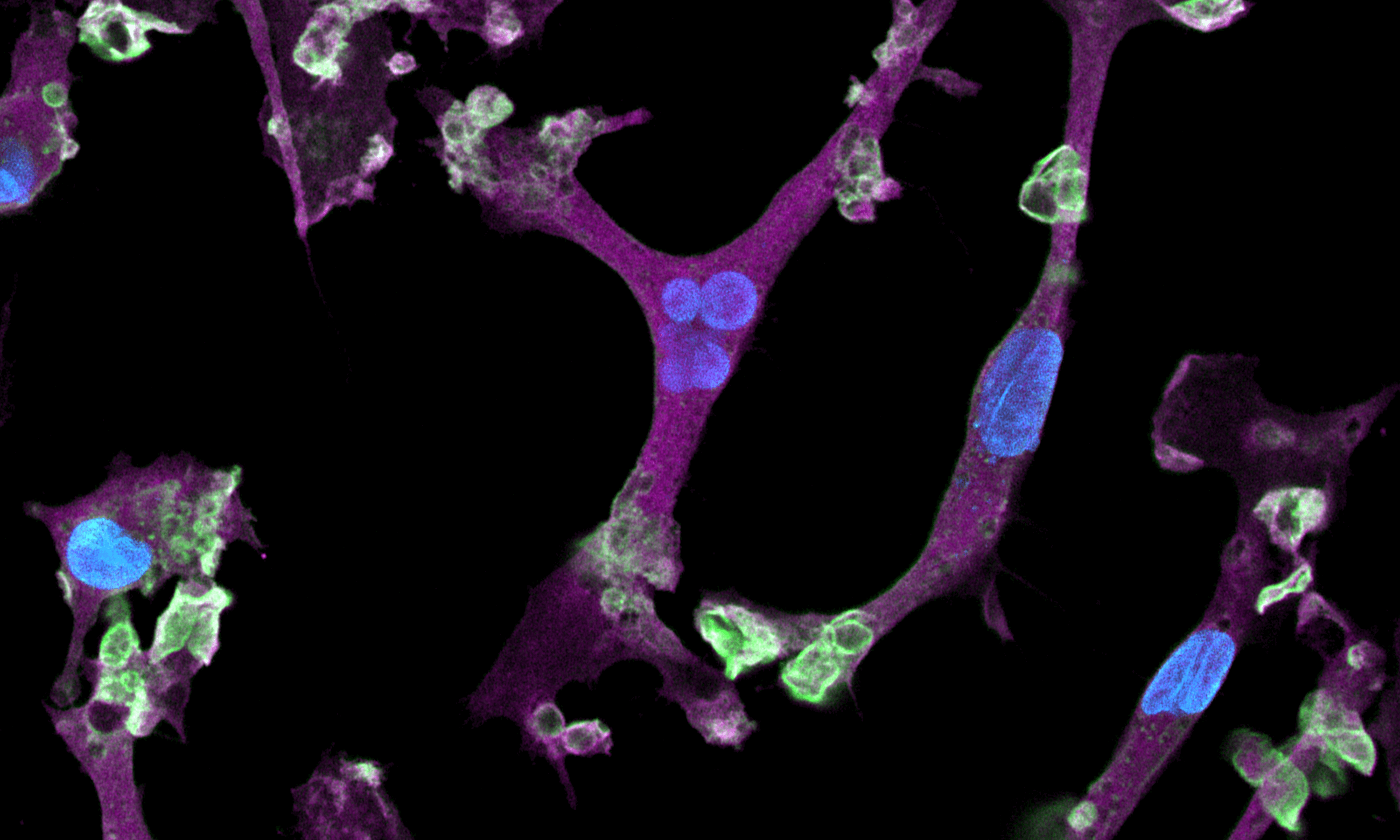
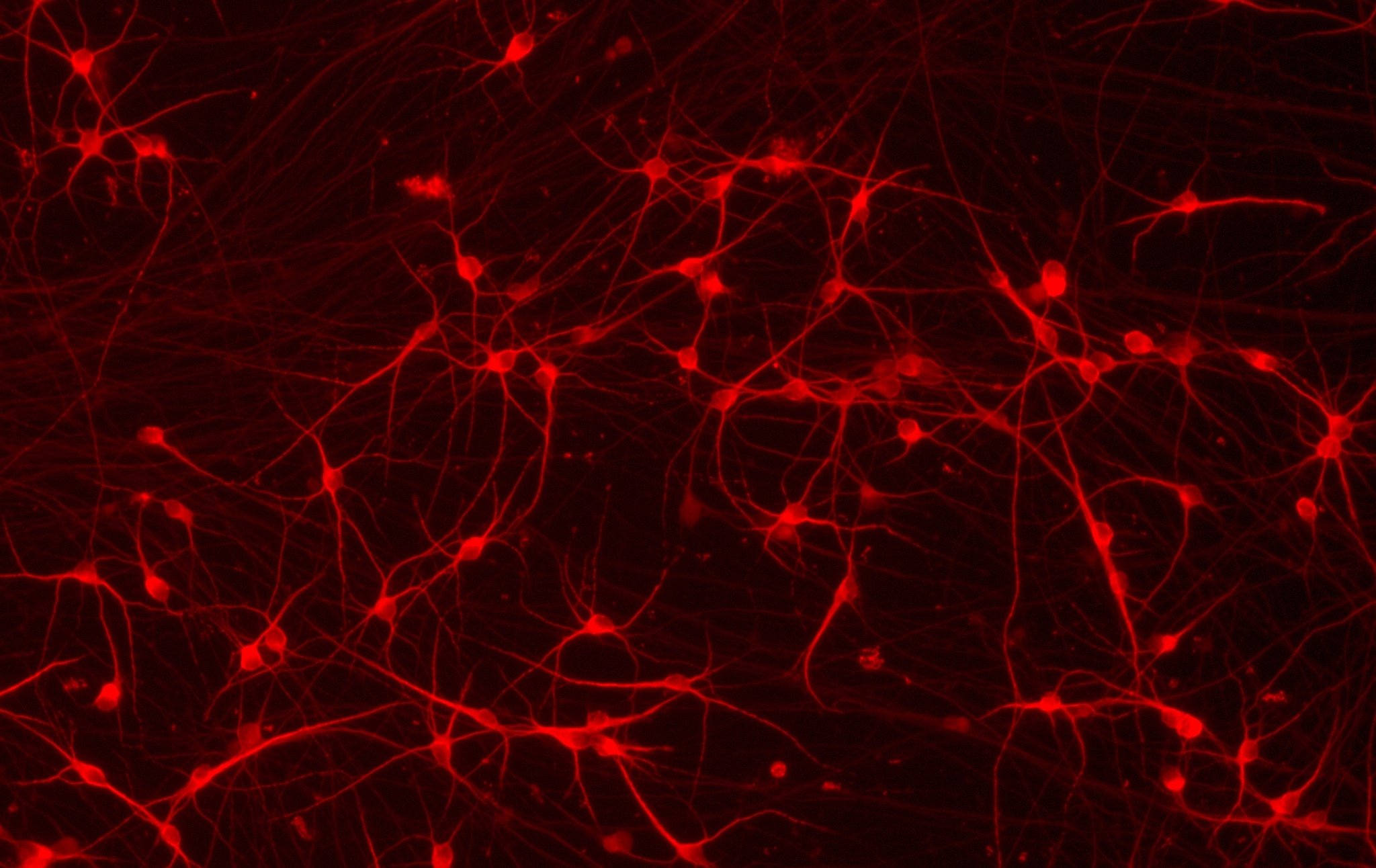
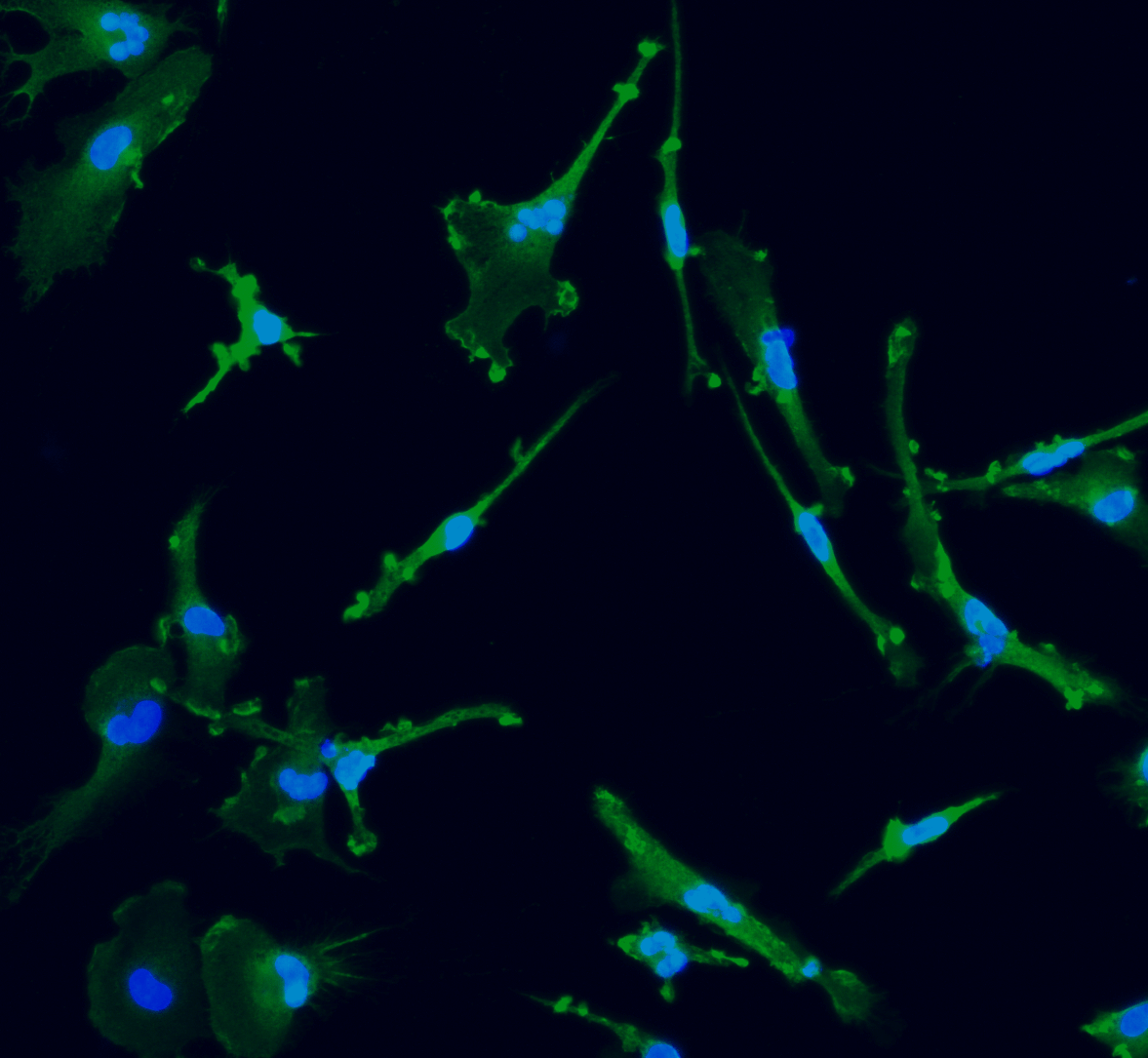
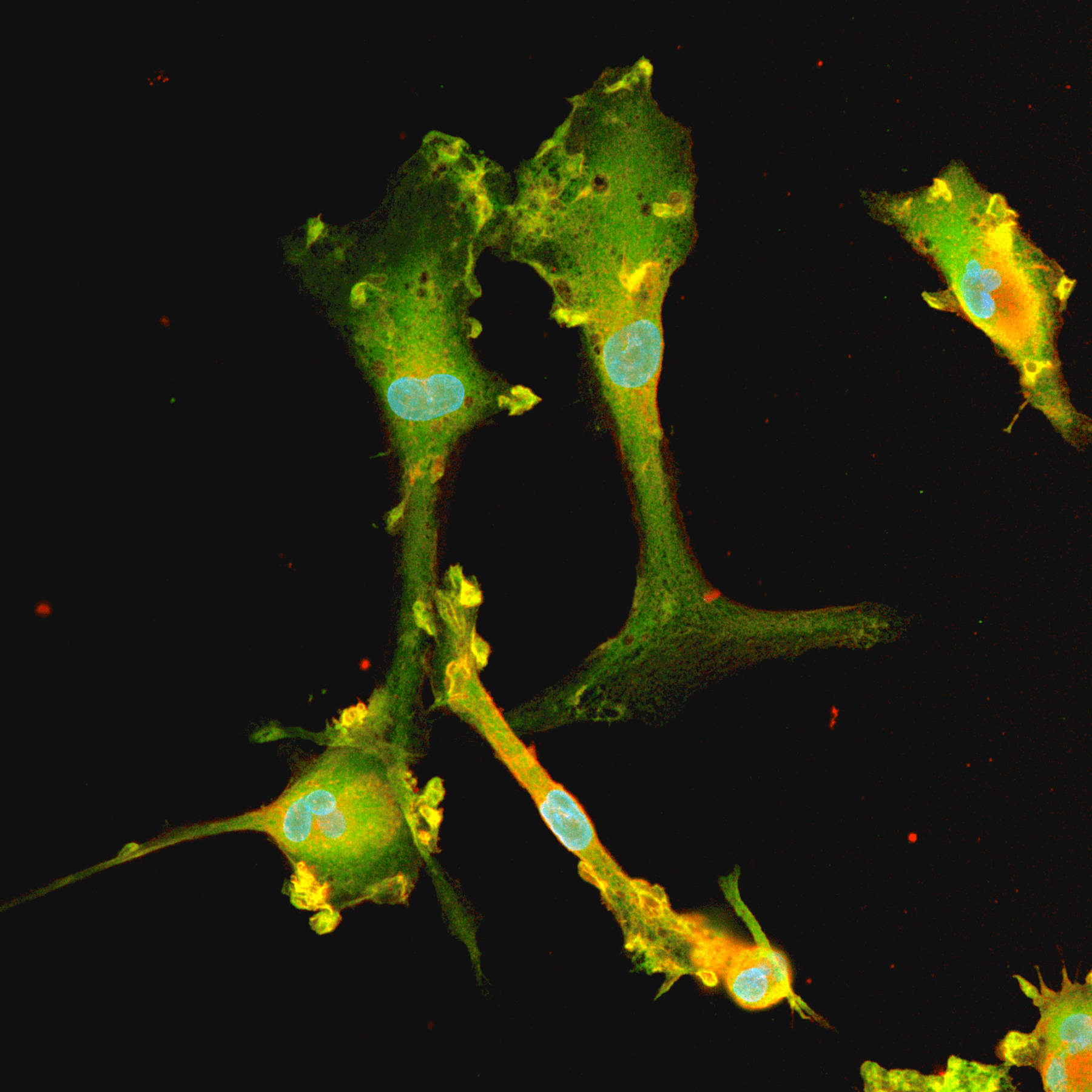
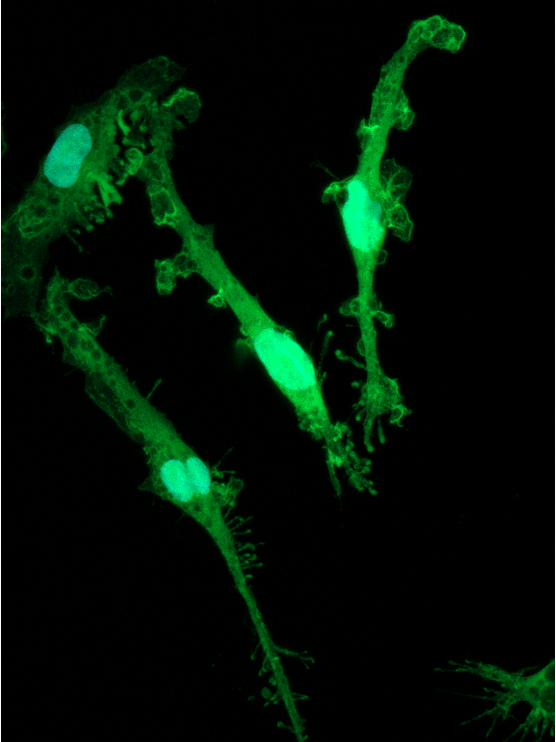
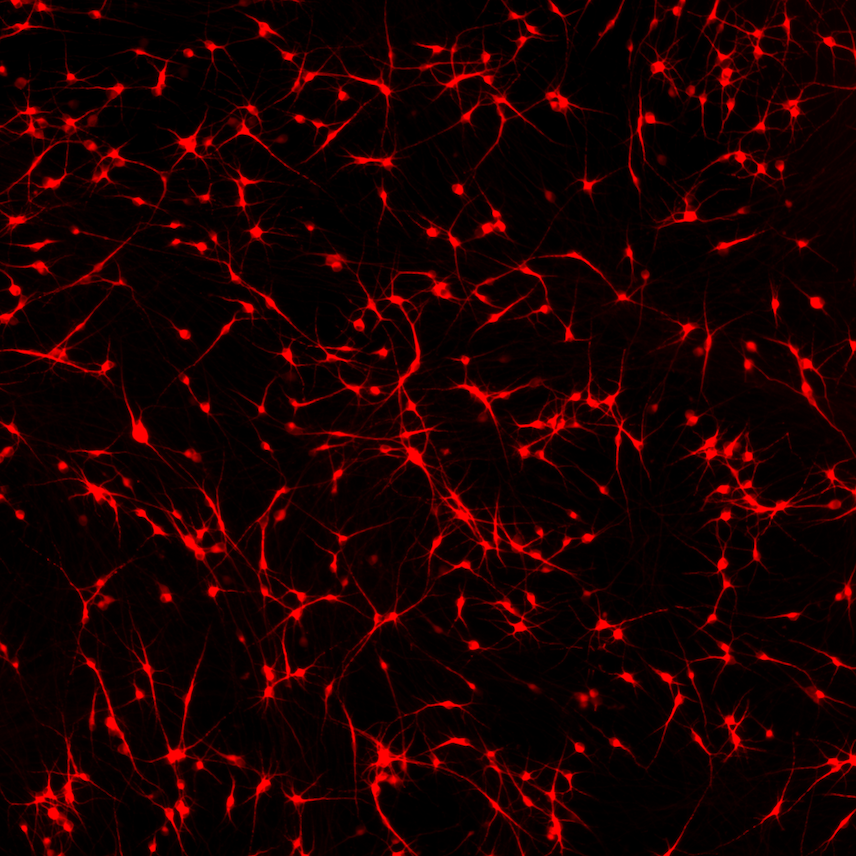
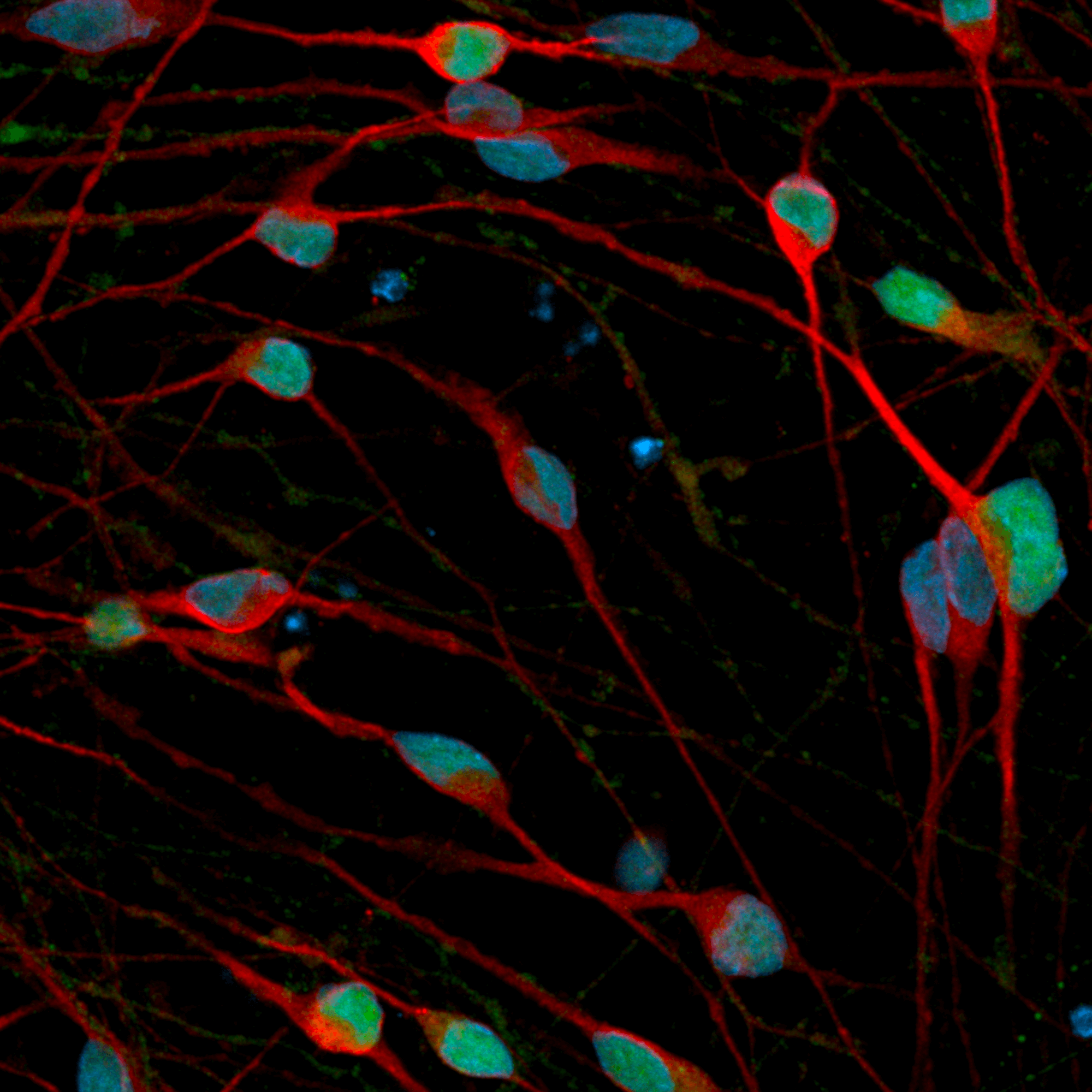
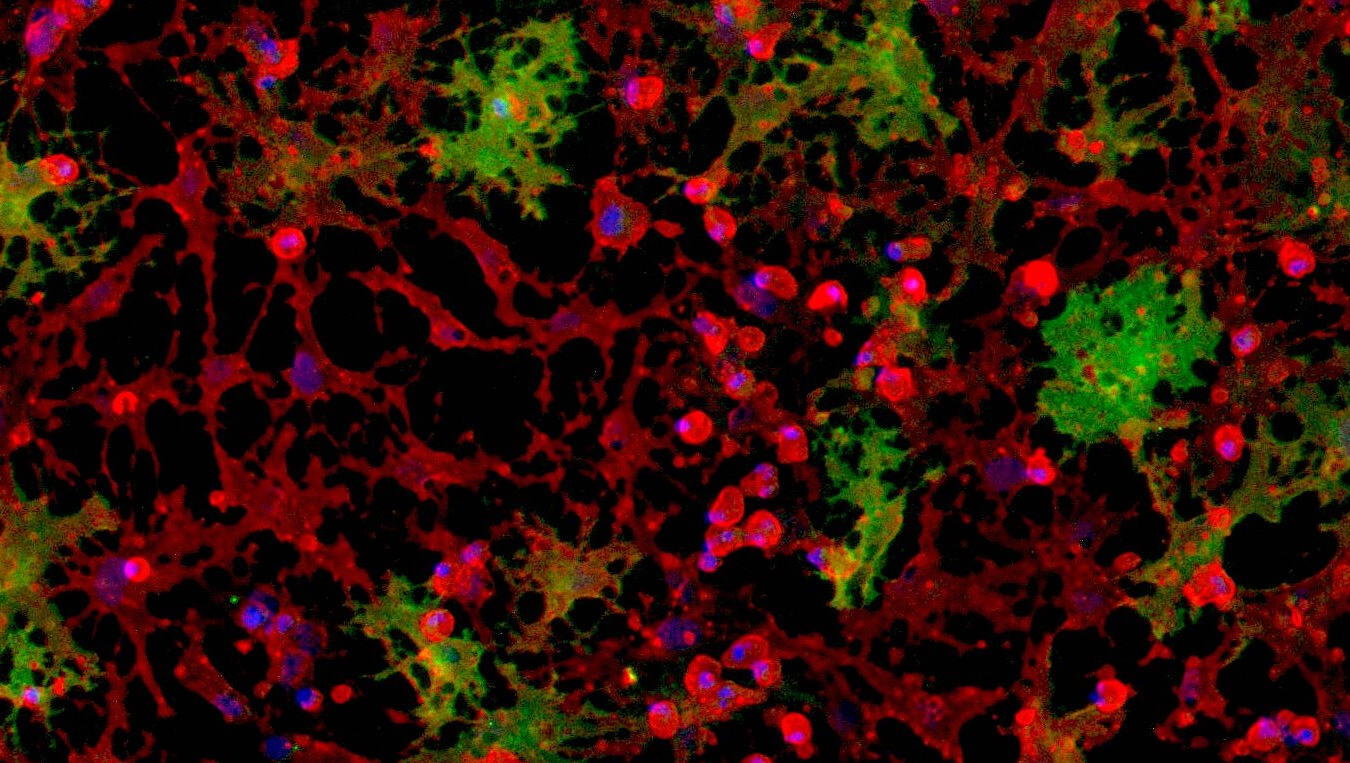
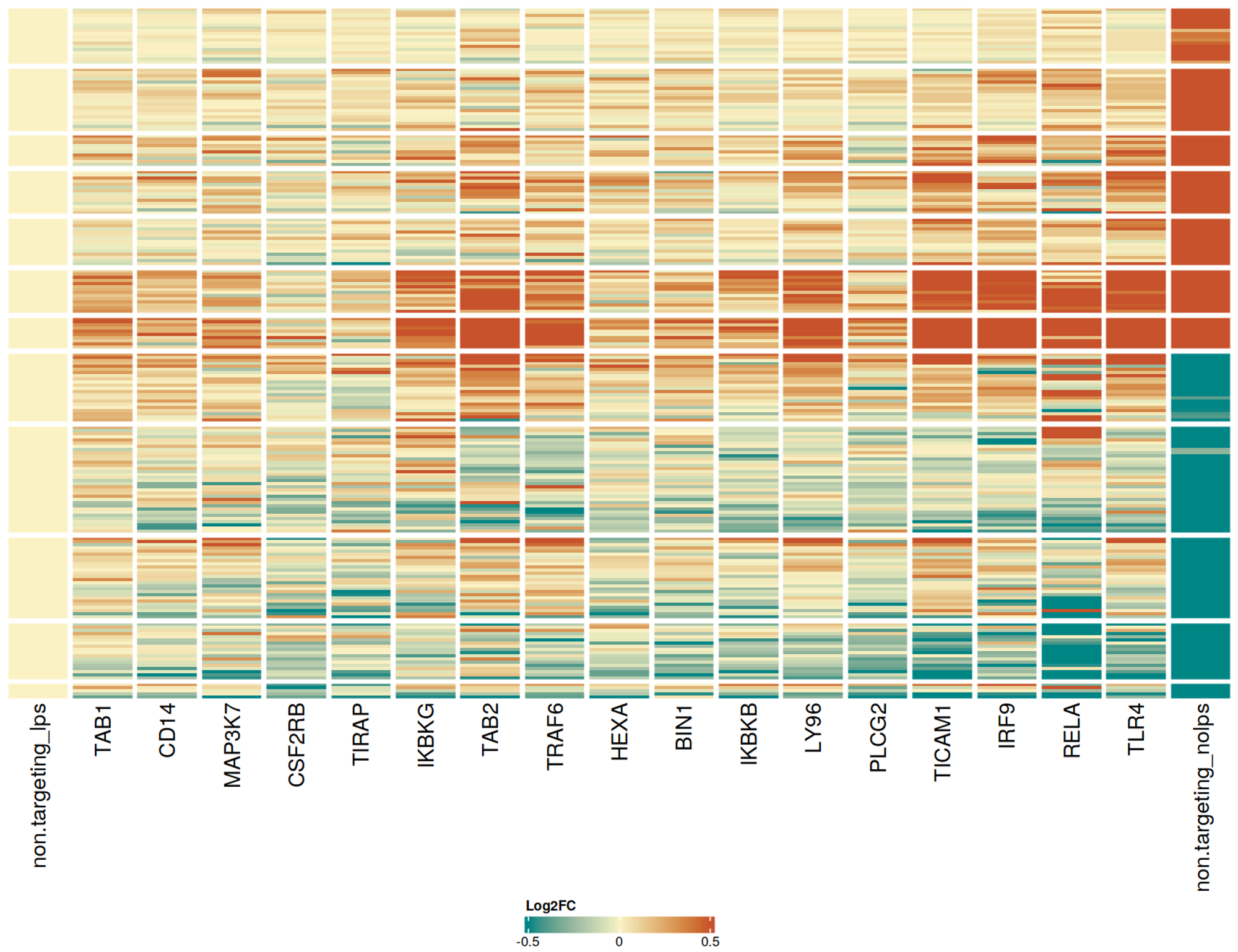
CRISPRko-Ready ioMicroglia are built from our well-established wild type ioMicroglia Male, engineered to constitutively express Cas9 nuclease. These cells arrive ready for guide RNA (gRNA) delivery from day 1 to 18 post-thaw. Using our optimised lentivirus or lipid-based gRNA delivery protocol, users can maximise their knockout efficiency and start measuring readouts from gene knockouts and CRISPR screens within days.
Our cells arrive ready to use for functional genomics, disease model generation, drug target identification and fundamental human biology research. The cells have been deterministically programmed from human iPSC using opti-ox technology, meaning scalability and consistency are built-in. In days, they convert consistently to microglia characterised by >90% expression of P2RY12 and IBA1.
Users can significantly cut experimental timelines by no longer needing to spend months engineering and characterising their own Cas9 stable iPSC lines or optimising differentiation protocols. With these cells, robust experimental readouts can be achieved by simply delivering gRNAs against your target gene. Users do not require prior expertise in iPSC differentiation or gRNA delivery optimisation.















 Emmanouil Metzakopian
Emmanouil Metzakopian