













cat no | io1007
ioGlutamatergic Neurons GBA null/R159W
Human iPSC-derived Gaucher and Parkinson’s disease model
-
Cryopreserved human iPSC-derived cells powered by opti-ox that are ready for experiments in days
-
Engineered to carry a GBA mutation relevant for modelling Gaucher and Parkinson's diseases
-
Consistent, functional excitatory neurons that form neuronal networks within days

Human iPSC-derived Gaucher and Parkinson’s disease model

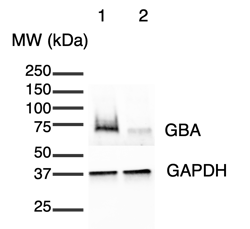
GBA protein is present in ioGlutamatergic Neurons GBA null/R159W at a lower level than the wild type control
A Western blot experiment confirmed the presence of the GBA protein in ioGlutamatergic Neurons GBA null/R159W at a lower level than in the wild-type ioGlutamatergic Neurons. Day 11 cell lysates were subjected to Western blotting (20 µg protein in 40 µl per lane) using 4-20% mini protean TGX stain-free gels. Proteins were transferred onto PVDF membranes using the Trans-Blot Turbo Transfer Pack, blocked for 10 minutes, incubated with primary antibodies (GBA Invitrogen MA5-26589, 1:2000; GAPDH Abcam ab8245, 1:5000), washed three times, incubated with HRP-labelled secondary antibodies, washed three times and signal visualised by electrochemiluminescence.
1= ioGlutamatergic Neurons (wild type), 2= ioGlutamatergic Neurons GBA null/R159W.

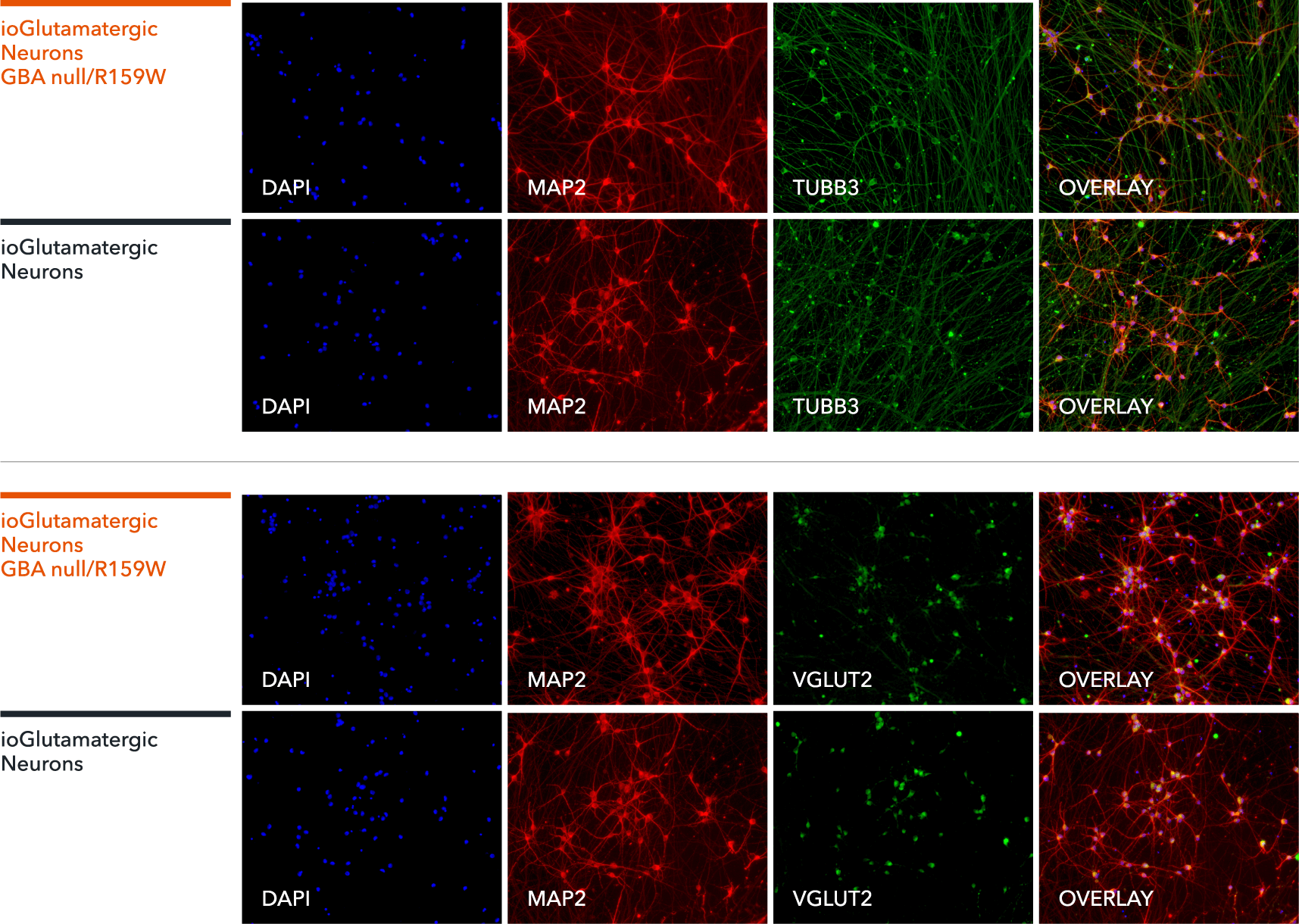
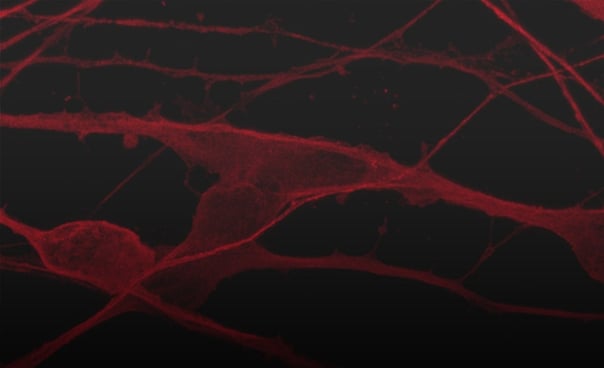
ioGlutamatergic Neurons GBA null/R159W express neuron-specific markers comparably to the isogenic control
Immunofluorescent staining on post-revival day 11 demonstrates similar homogenous expression of pan-neuronal proteins TUBB3 and MAP2 (upper panel) and glutamatergic neuron-specific transporter VGLUT2 (lower panel) in ioGlutamatergic Neurons GBA null/R159W compared to the isogenic control. 100X magnification.

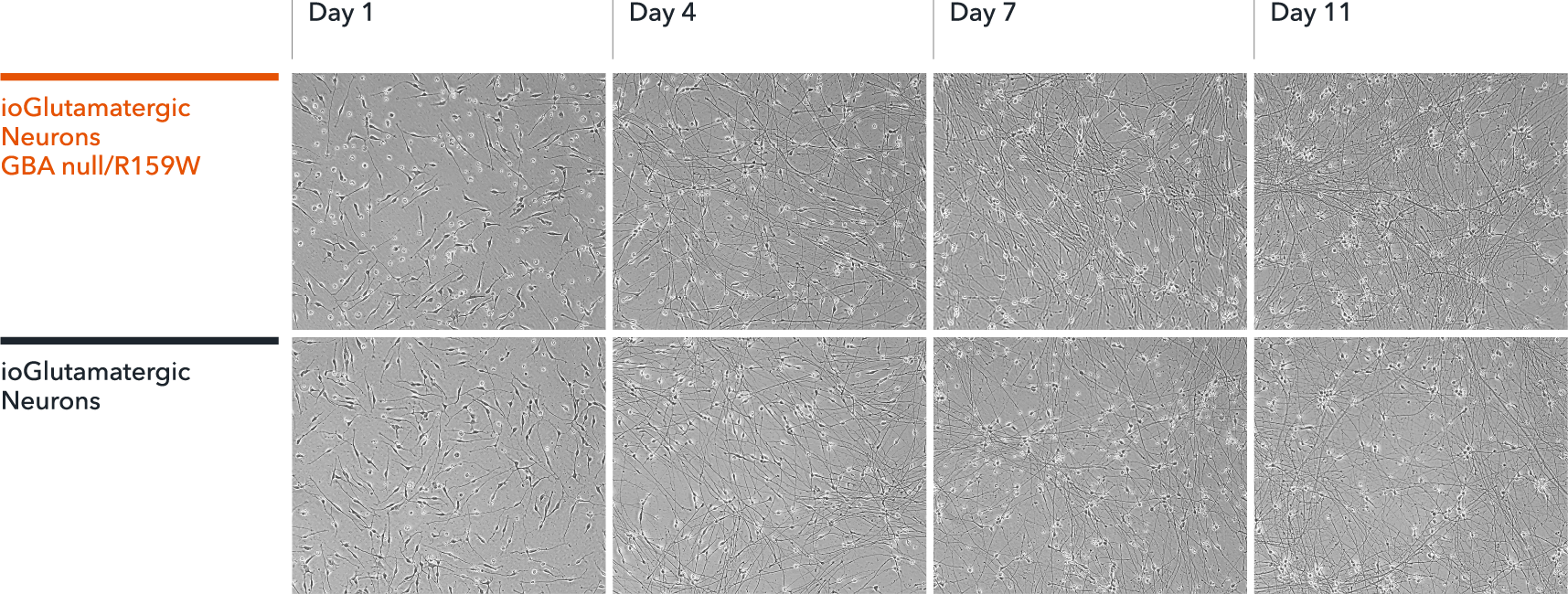
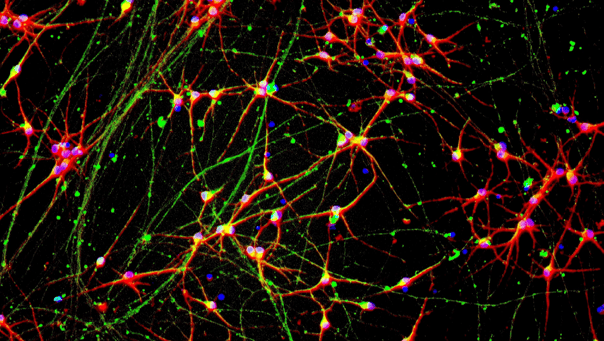
ioGlutamatergic Neurons GBA null/R159W form structural neuronal networks by day 11
ioGlutamatergic Neurons GBA null/R159W mature rapidly and form structural neuronal networks over 11 days, when compared to the isogenic control. Day 1 to 11 post thawing; 100X magnification.

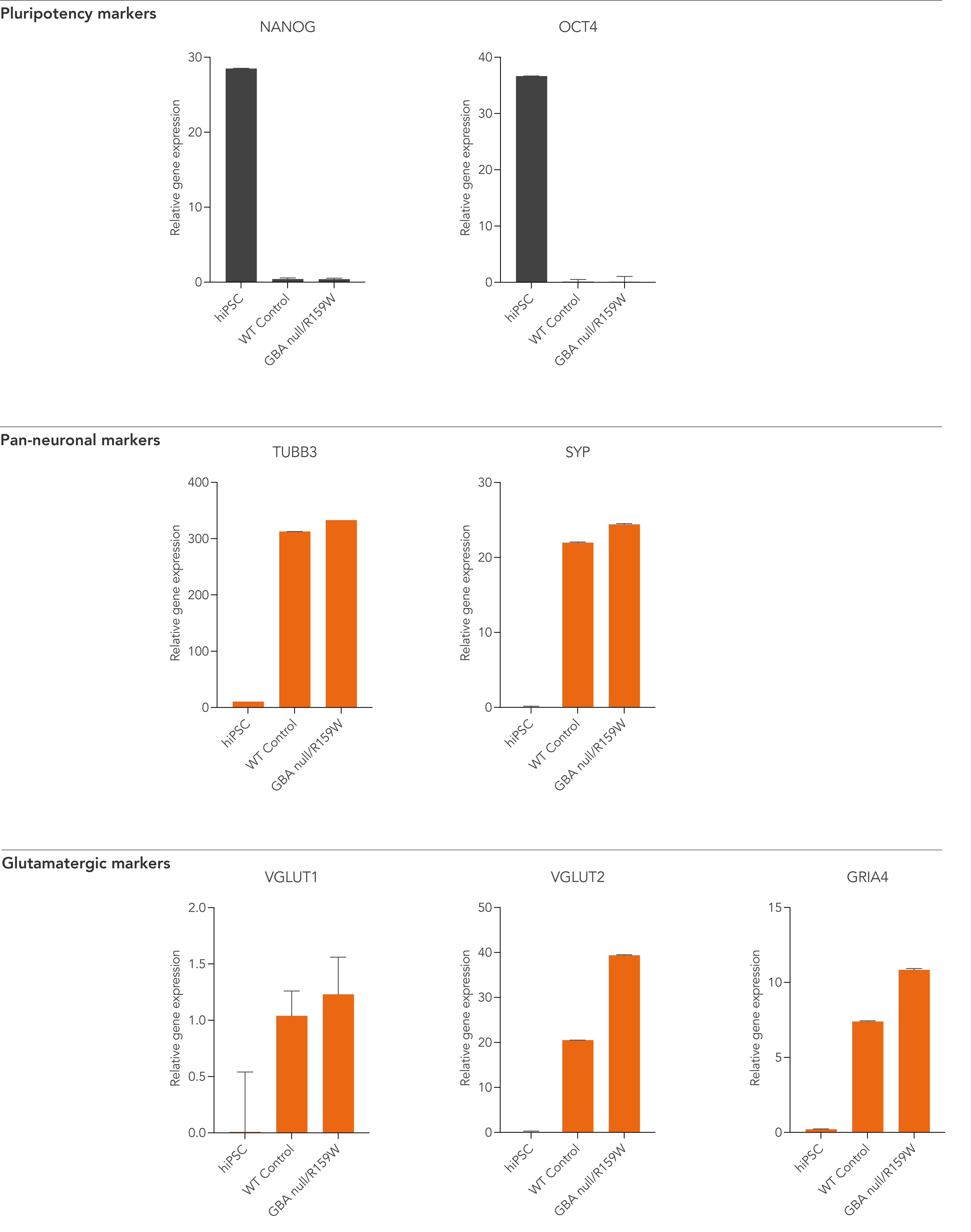
ioGlutamatergic Neurons GBA null/R159W demonstrate gene expression of neuronal and glutamatergic-specific markers following deterministic programming
Gene expression analysis demonstrates that ioGlutamatergic Neurons GBA null/R159W and the isogenic control (WT Control) lack the expression of pluripotency markers (NANOG and OCT4) at day 11, whilst robustly expressing pan-neuronal (TUBB3 and SYP) and glutamatergic specific (VGLUT1 and VGLUT2) markers, as well as the glutamate receptor GRIA4. Gene expression levels were assessed by RT-qPCR (data normalised to HMBS; cDNA samples of the parental human iPSC line (hiPSC) were included as reference). Data represents day 11 post-revival samples, n=2 replicates.

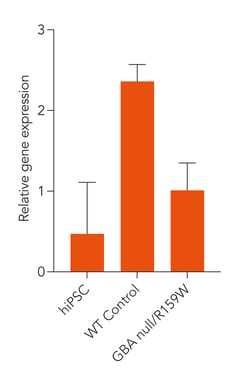
Disease-related GBA is expressed in ioGlutamatergic Neurons GBA null/R159W following deterministic programming
Gene expression analysis demonstrates that ioGlutamatergic Neurons GBA null/R159W and the isogenic control (WT Control) express the GBA gene encoding the glucocerebrosidase protein. Gene expression levels were assessed by RT-qPCR (data normalised to HMBS, cDNA samples of the parental human iPSC line (hiPSC) were included as reference). Data represents day 11 post-revival samples, n=2 replicates.

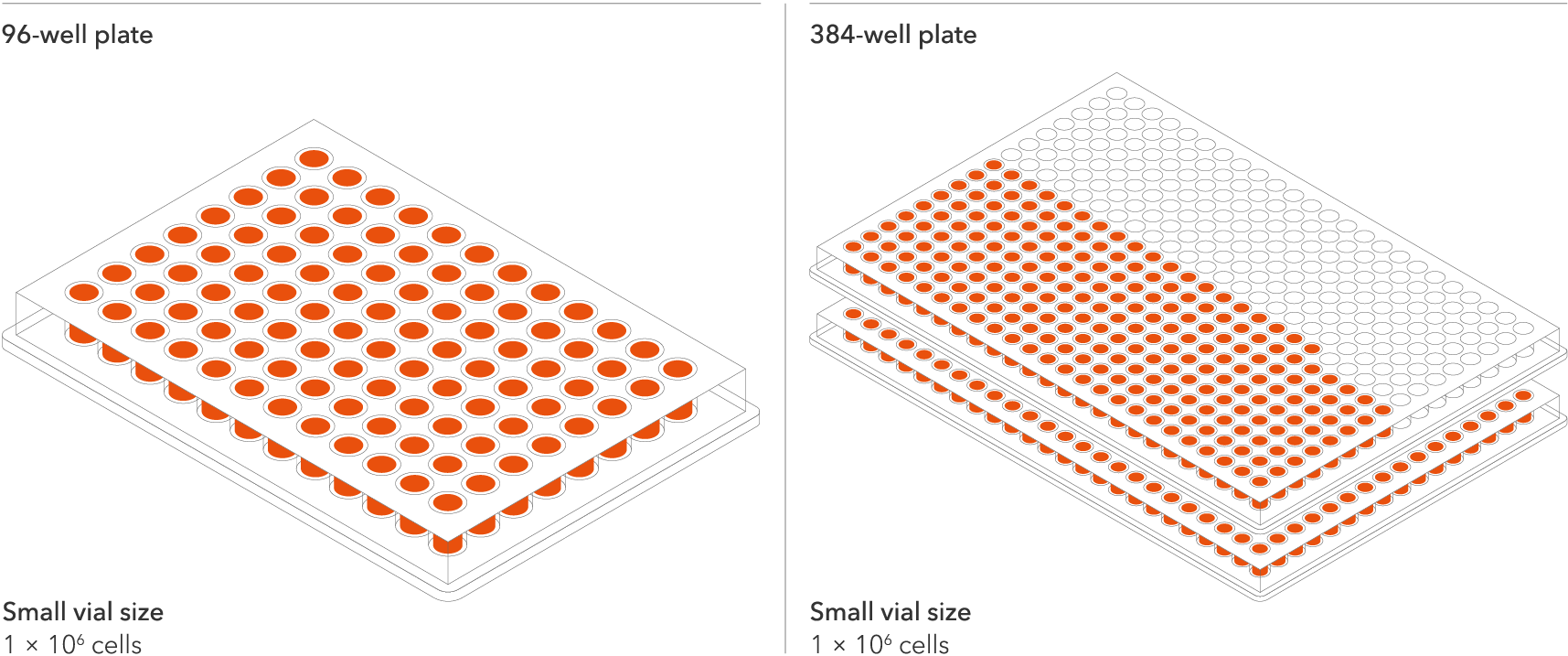
Industry leading seeding density
The recommended minimum seeding density is 30,000 cells/cm2, compared to up to 250,000 cells/cm2 for other similar commercially available products. One small vial can plate a minimum of 0.7 x 24-well plate, 1 x 96-well plate, or 1.5 x 384-well plates. This means every vial goes further, enabling more experimental conditions and more repeats, resulting in more confidence in the data.
Vial limit exceeded
A maximum number of 20 vials applies. If you would like to order more than 20 vials, please contact us at orders@bit.bio.




Hoescht(blue)_day12v2.png?width=604&name=bit.bio_ioGlutamatergic%20Neurons_20xMAP2(red)Hoescht(blue)_day12v2.png)
Hoescht(blue)TUBB3(blue)_day4.jpg?width=604&name=bit.bio_ioGlutamatergic%20Neurons_60xMAP2(red)Hoescht(blue)TUBB3(blue)_day4.jpg)

.png?width=1860&height=1260&name=bit.bio_3x2_ioGlutamatergic%20Neurons_MAP2_Hoescht_x20_hi.res%20(1).png)









